Slot And Dot Blot Method
Dot-Blot method description What solution can be applied in a dot? Protein Total protein is isolated in 1x PEB buffer prepared freshly from 4x stock (AGRISERA, AS08 300) according to Agrisera protocol (protease inhibitor cocktail must be included). Protein amounts are determined using 1/5 of the volumes of the standard DC Protein assay (BIORAD) against a BGG standard dilution curve of 0.0, 0.2. A Dot Blot is a simple and quick assay that may be employed to determine if your antibodies and detection system are effective. Dot Blot may also be used to determine appropriate starting concentration of primary antibody for Western blot. Use a strip of nitrocellulose membrane. A Southern Blot is used to detect the presence of a certain sequence of DNA in your sample (Western Blot= proteins, Northern= RNA). Similarly, a slot blot is a method for detecting specific DNA, RNA or proteins in your sample. The key difference i. Dot-Blot method description What solution can be applied in a dot? Protein Total protein is isolated in 1x PEB buffer prepared freshly from 4x stock (AGRISERA, AS08 300) according to Agrisera protocol (protease inhibitor cocktail must be included). Protein amounts are determined using 1/5 of the volumes of the standard DC Protein assay (BIORAD) against a BGG standard dilution curve of 0.0, 0.2.
^ 'Peter Jackson's King Kong'.: 101. Retrieved 2014-01-05. Kong the 8th wonder of the world. Pavlovic, Uros 'Vader' (2005-11-25).
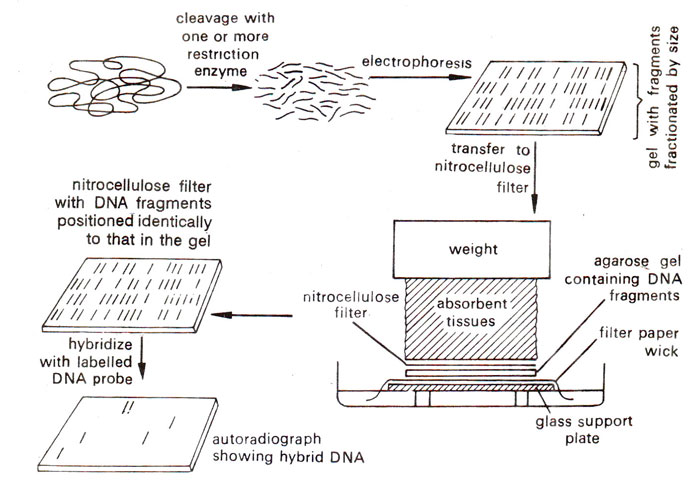
Dot Blot Protocol
- Barinaga, M., Franco, R., Meinkoth, J., Ong, E. and Wahl, G.M. (1981) Methods for the Transfer of DNA, RNA and Protein to Nitrocellulose and Diazotized Paper Solid Supports. A Schleicher and Scheull ManualGoogle Scholar
- Bio-Rad Laboratories Manual (1983) Bio-Dot Microfiltration Apparatus Instruction ManualGoogle Scholar
- Bresser, J. and Gillespie, D. (1983) Quantitative Binding of Covalently Closed Circular DNA to Nitrocellulose in Nal, Anal. Biochem., 129, 357–364CrossRefGoogle Scholar

References
- Boffey, S.A. (1984) Plasmid DNA Isolation by the Cleared Lysate Method, in Walker, J.M. (ed.) Methods in Molecular Biology, Volume 2, pp. 177–184. (Humana Press, New Jersey)Google Scholar
- Dale, J.W. and Greenaway, P.J. (1984) Plasmid DNA Isolation (Sheared Lysate Method), in Walker, J.M. (ed.) Methods in Molecular Biology, Volume 2, pp. 185–190. (Humana Press, New Jersey)Google Scholar
- Dale, J.W. and Greenaway, P.J. (1984) Preparation of Phage Lambda DNA, in Walker, J.M. (ed.) Methods in Molecular Biology, Volume 2, pp. 211–216. (Humana Press, New Jersey)Google Scholar
- Mathew, C.G.P. (1984) The Isolation of High Molecular Weight Eukaryotic DNA, in Walker, J.M. (ed.) Methods in Molecular Biology, Volume 2, pp. 31–34. (Humana Press, New Jersey)Google Scholar
- Kafatos, F.C., Jones, C.W. and Efstratiadis, A. (1979) Determination of Nucleic Acid Sequence Homologies and Relative Concentrations by a Dot Hybridization Procedure, Nucleic Acids Res., 7, 1541–1552CrossRefGoogle Scholar
- Thomas, P.S. (1979) Hybridization of Denatured RNA and Small DNA Fragments Transferred to Nitrocellulose, Proc. Natl. Acad. Sci. USA, 77, 5201–5205CrossRefGoogle Scholar
- Slater, R.J. (1983) The Extraction and Fractionation of RNA, in Walker, J.M. and Gaastra, W. (eds) Techniques in Molecular Biology, Volume 1, pp. 113–134. (Croom Helm, London/Macmillan, New York)Google Scholar
- Cheley, S. and Anderson, R. (1984) A Reproducible Microanalytical Method for the Detection of Specific RNA Sequences by Dot-Blot Hybridization, Anal. Biochem., 137, 15–19CrossRefGoogle Scholar
- Thomas, P.S. (1983) Hybridization of Denatured RNA Transferred or Dotted to Nitrocellulose Paper, Methods in Enzymology, 100, pp. 255–266. (Academic Press, London)Google Scholar
- Thiele, C.J., Reynolds, C.P. and Israel, M.A. (1985) Decreased Expression of N-myc Precedes Retinoic Acid-induced Morphological Differentiation of Human Neuroblastoma, Nature, Lond., 313, 404–406CrossRefGoogle Scholar
- Hamaguchi, K. and Geiduschek, E.P. (1962) The Effect of Electrolytes on the Stability of the Deoxyribonucleate Helix, J. Amer. Chem. Soc., 84, 1329–1338CrossRefGoogle Scholar
- Manser, T. and Gefter, M.L. (1984) Isolation of Hybridomas Expressing a Specific Heavy Chain Variable Region Gene Segment by Using a Screening Technique that Detects mRNA Sequences in Whole Cell Lysates, Proc. Natl. Acad. Sci. USA, 81, 2470–2474CrossRefGoogle Scholar
- Bresser, J., Hubbell, H.R. and Gillespie, D. (1983) Biological Activity of mRNA Immobilized on Nitrocellulose in Nal, Proc. Natl. Acad. Sci. USA, 80, 6523–6527CrossRefGoogle Scholar
- Gillespie, D., Caranfa, M.J. and Bresser, J. (1984) The Role of Chaotropic Salts in Two-phase Gene Diagnosis, BioEssays, 1, 272–276CrossRefGoogle Scholar
- Mathew, C.G.P. (1983) Labelling DNA in vitro — Nick Translation, in Walker, J.M. and Gaastra, W. (eds) Techniques in Molecular Biology, pp. 159–166. (Croom Helm, London/Macmillan, New York)Google Scholar
- Mathew, C.G.P. (1983) Detection of Specific DNA Sequences — The Southern Blot, in Walker, J.M. and Gaastra, W. (eds) Techniques in Molecular Biology, pp. 273–286. (Croom Helm, London/Macmillan, New York)Google Scholar
- Leary, J.J., Brigati, D.J. and Ward, D.C. (1983) Rapid and Calorimetric Method for Visualizing Biotin-labelled DNA Probes Hybridized to DNA or RNA Immobilized on Nitrocellulose: Bio-blots, Proc. Natl. Acad. Sci. USA, 80, 4045–4049CrossRefGoogle Scholar
- Hawkes, R., Niday, E. and Gordon, J. (1982) A Dot-immunobinding Assay for Monoclonal and Other Antibodies, Anal. Biochem., 119, 142–147CrossRefGoogle Scholar
- Renart, J., Reiser, J. and Stark, G.R. (1979) Transfer of Small DNA Fragments from Polyacrylamide Gels to Diazobenzyloxymethyl-paper and Detection by Hybridization with DNA Probes, Biochem. Biophys. Res. Comm., 85, 1104–1112Google Scholar
- Stark, G.R. and Wahl, G.M. (1984) Gene Amplification, Ann. Rev. Biochem., 53, 447–491CrossRefGoogle Scholar
- Griffen-Shea, R., Thireos, G., Kafatos, F.C., Petri, W.H. and Villa-Komaroff, C. (1980) Chorion cDNA Clones of D. melanogaster and Their Use in Studies of Sequence Homology and Chromosomal Location of Chorion Genes, Cell, 19, 915–922CrossRefGoogle Scholar
- Dalgleish, R., Trapnell, B.C., Crystal, R.G. and Tolstoshev, P. (1982) Copy Number of a Human Type 1 a 2 Collagen Gene, J. Biol. Chem., 257, 13816–13822Google Scholar
- Yaswen, P., Goyette, M., Shank, P.R. and Fausto, N. (1985) Expression of c-Ki-ras, c-Ha-ras and c-myc in Specific Cell Types During Hepatocarcinogenesis, Molec. Cell. Biol., 5, 780–786Google Scholar
- Reitsma, P.H., Rothberg, P.H., Astrin, S.M., Trial, J., Bar-Sharit, Z., Hall, A., Teitelbaum, S.L. and Kahn, A.J. (1983) Regulation of myc Gene Expression in HL-60 Leukemia Cells by a Vitamin D Metabolite, Nature, Lond., 306, 492–494CrossRefGoogle Scholar
- Greenberg, M.E. and Ziff, E.B. (1984) Stimulation of 3T3 Cells Induces Transcription of the C-fos Proto-oncogene, Nature, Lond, 311, 433–438CrossRefGoogle Scholar
Dot Blot Method
Dot blot technique is a method of identifying the presence of DNA, RNA and Protein in the sample. It is the simplified form of Southern, Northern and Western blotting for the isolation of DNA, RNA and protein respectively. This method can detect the presence or absence of biomolecule in a single run. Dot blot technique is a very popular method. In this dot blot approach, a gDNA mixture containing the methylated DNA to be detected was spotted directly onto an N + membrane as a dot inside a previously drawn circular template pattern. Compared with other gel electrophoresis-based blotting approaches and other complex blotting procedures, the dot blot method saves significant time.